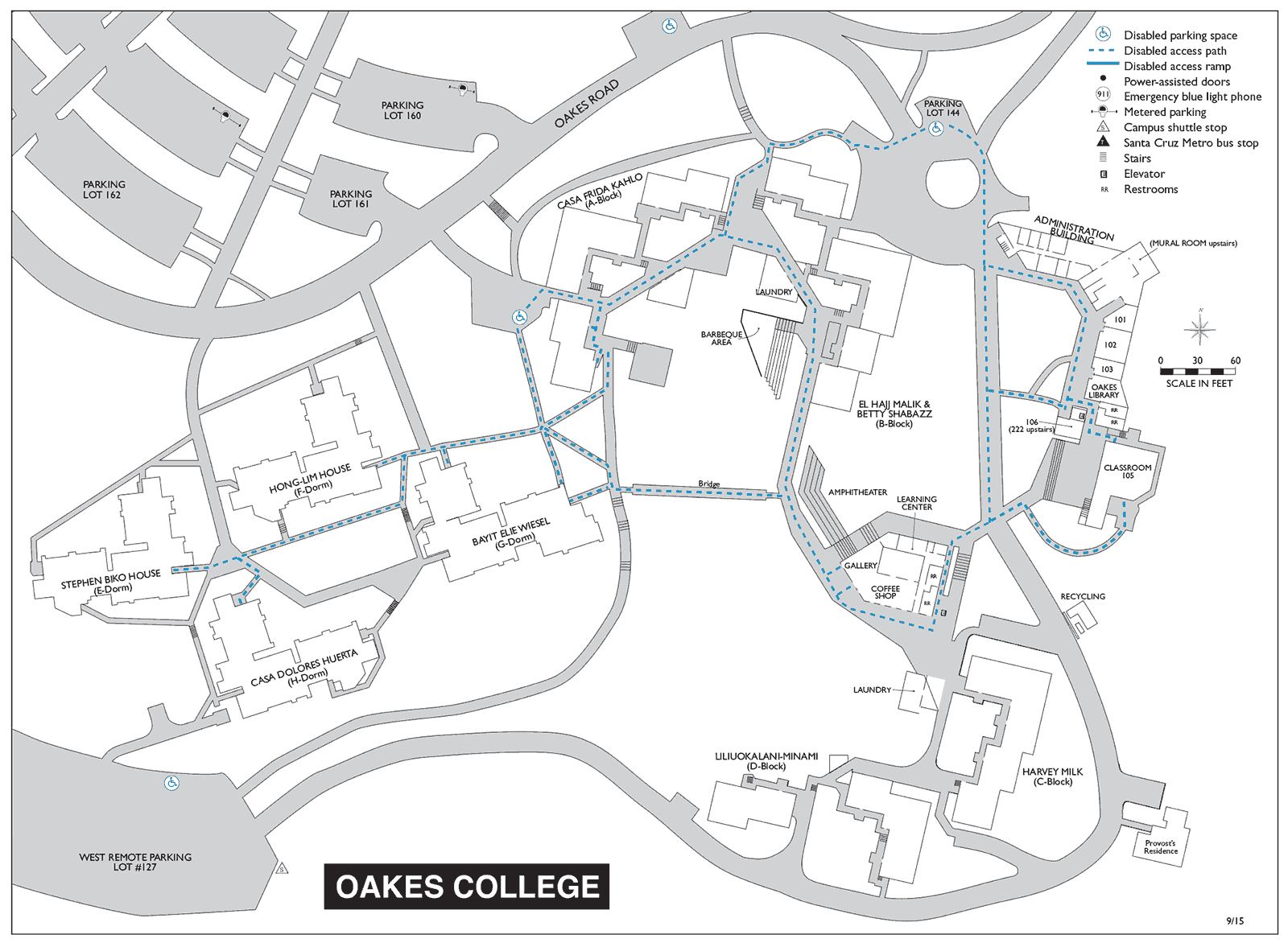
gneiss.plot._blobtree — gneiss 0.2 documentation The jkruppa/virDisco package contains the following man pages: aa_info_db aa_seqs artificial_genome_mapping build_index build_sample_info check_host collapse_df coverage_filter dna_seqs empty_map_pep_list ete2_plot fastq_quality_control fastq_trimmer fq2fa generate_decoy_reads generate_read get_bowtie2_cmd get_consensus_df get_pauda_hits get
Phylogenetic Course Exercises — Introduction to
The Programmable Tree Drawing Engine — ETE Toolkit. ete3 [options] In the real submission script, at least all the above underlined values need to be reviewed or to be replaced by the proper values. Please refer to Running Jobs on Sapelo2 , Run X window Jobs and Run interactive Jobs for more details about running jobs at Sapelo2., Overview¶ This package focuses on inferring orthologs using NCBI’s blast, various sequence alignment strategies, and phylogenetics analyses including PAML, PhyML, ete3, and more tools..
30.10.2019В В· Learn about Jupyter Notebooks and how you can use them to run your code. . Codecademy is the easiest way to learn how to code. It's interactive, fun, and you can do it with your friends. ete3 [options] In the real submission script, at least all the above underlined values need to be reviewed or to be replaced by the proper values. Please refer to Running Jobs on Sapelo2 , Run X window Jobs and Run interactive Jobs for more details about running jobs at Sapelo2.
27.11.2018В В· Tools for working with the NCBI taxonomy database (and DIAMOND output) - 0.1.1 - a Python package on PyPI - Libraries.io *** By default, the R Studio session information will be stored in your home directory. If you are working with big data files, these files could be very big and fill up your home directory ***
grimm.py¶ class bg.grimm.GRIMMReader ¶ Bases: object. Class providing a staticmethod based implementation of reading GRIMM formatted data file-like object and obtain a bg.breakp Features. Individual based model of evolving microbes. Explicit model of evolving genome. Spatially extended environment. Microbes evolve metabolic capacities to exploit environmental nutrients.
Source code for gneiss.balances""" Balances (:mod:`gneiss.balances`) =====.. currentmodule:: gneiss.balances This module contains modules for calculating balances and creating ETE objects to visualize these balances on a tree. Overview ¶ Phylogenetic trees are the result of most evolutionary analyses. They represent the evolutionary relationships among a set of species or, in molecular biology, a …
Phylogenetic Course: Exercises¶ Basic phylogenetic tree making. We are going to use this to highlight some basic concepts and few exercises to learn tree making using some popular methods. Later we will use the workflows via ete toolkit version 3 or ete3 Introduction to Phylogenetics¶ Contents¶ Reasons for making a phylogenetic tree. Molecular phylogenetics
ete3 [options] In the real submission script, at least all the above underlined values need to be reviewed or to be replaced by the proper values. Please refer to Running Jobs on Sapelo2 , Run X window Jobs and Run interactive Jobs for more details about running jobs at Sapelo2. ete3 website ETE is a python programming toolkit that assists in the automated manipulation, analysis and visualization of phylogenetic trees. It provides a wide range of tree handling options, node annotation features, programmatic access to the phylomeDB database (containing thousands of pre-calculated phylogenetic trees), and automatic orthology and paralogy detection.
The toytree ethos¶ Welcome to toytree, a minimalist tree manipulation and plotting library. Toytree combines a popular Tree data structure based on the ete3 library with modern plotting tools based on the toyplot plotting library. However, for power users, the TreeNode structure of toytrees provides a lot of additional functionality especially for doing scientific computation and research on trees. The TreeNode object in toytree is a modified fork of the TreeNode in ete3. Thus, you can read the very detailed ete documentation if you want a detailed understanding of the
conda install noarch v3.1.1; To install this package with conda run: conda install -c etetoolkit ete3 Funannotate Documentation, Release 1.0.1 docker build -t funannotate -f Dockerfile . Running the Docker container with your data: In order to run the docker container, you need to put all the п¬Ѓles you will use for input to funannotate into the same
Source code for gneiss.balances""" Balances (:mod:`gneiss.balances`) =====.. currentmodule:: gneiss.balances This module contains modules for calculating balances and creating ETE objects to visualize these balances on a tree. 30.10.2019В В· Learn about Jupyter Notebooks and how you can use them to run your code. . Codecademy is the easiest way to learn how to code. It's interactive, fun, and you can do it with your friends.
3. Installing ProPhyle¶ ProPhyle is written in Python, C and C++. It is distributed as a Python package and all C/C++ programs are compiled upon the first execution of the main program. Phylogenetic Course: Exercises¶ Basic phylogenetic tree making. We are going to use this to highlight some basic concepts and few exercises to learn tree making using some popular methods. Later we will use the workflows via ete toolkit version 3 or ete3
phyphy documentation — phyphy documentation. The toytree ethos¶ Welcome to toytree, a minimalist tree manipulation and plotting library. Toytree combines a popular Tree data structure based on the ete3 library with modern plotting tools based on the toyplot plotting library., Finally we prepared TreeMatcher for future merge with it's parent project (ETE3) and the documentation was updated. (link 3) Interesting parts . The categorization of metacharacters (period 1) The ability to use pure Python expressions to define a node (period 2) Limitations are met at describing nodes' descendants relax patterns (period2).
Tomas Kovalovsky Configuration management section leader
from_pandas_dataframe — NetworkX 1.10 documentation. 27.11.2018 · Tools for working with the NCBI taxonomy database (and DIAMOND output) - 0.1.1 - a Python package on PyPI - Libraries.io, 3. Installing ProPhyle¶ ProPhyle is written in Python, C and C++. It is distributed as a Python package and all C/C++ programs are compiled upon the first execution of the main program..
pyham В· PyPI
GitHub etetoolkit/ete Python package for building. 10.05.2018В В· GitHub is home to over 40 million developers working together to host and review code, manage projects, and build software together *** By default, the R Studio session information will be stored in your home directory. If you are working with big data files, these files could be very big and fill up your home directory ***.
Dependencies¶ Funannotate has a lot of dependencies. However, it also comes with a few tools to help you get everything installed. The first is that of funannotate check. Dependencies¶ Funannotate has a lot of dependencies. However, it also comes with a few tools to help you get everything installed. The first is that of funannotate check.
16.07.2018 · I read the whole documentation on profile face heatmap generation but I could not find any way to add labels to the heatmap produced by ete3. For … Several types of node faces are provided by the main ete3 module, ranging from simple text (TextFace) and geometric shapes (CircleFace), to molecular sequence representations (SequenceFace), heatmaps and profile plots (ProfileFace). A complete list of available faces can …
The toytree ethos¶ Welcome to toytree, a minimalist tree manipulation and plotting library. Toytree combines a popular Tree data structure based on the ete3 library with modern plotting tools based on the toyplot plotting library. ipyrad-analysis toolkit: abba-baba¶ The baba tool can be used to measure abba-baba statistics across many different hypotheses on a tree, to easily group individuals into populations for measuring abba-baba using allele frequencies, and to summarize or plot the results of many analyses.
This will get you a list of ete3.TreeNode objects that represent the 5 most diverse possible strains. You can then use strainchoosr.get_leaf_names_from_nodes(diverse_strains) to get a list of names. Complete documentation on the strainchoosr API can be found at https://strainchoosr.readthedocs.io/api.html. Issues and Pull Requests However, for power users, the TreeNode structure of toytrees provides a lot of additional functionality especially for doing scientific computation and research on trees. The TreeNode object in toytree is a modified fork of the TreeNode in ete3. Thus, you can read the very detailed ete documentation if you want a detailed understanding of the
Phylogenetic Course: Exercises¶ Basic phylogenetic tree making. We are going to use this to highlight some basic concepts and few exercises to learn tree making using some popular methods. Later we will use the workflows via ete toolkit version 3 or ete3 Source code for aiida.cmdline.commands.work. # -*- coding: utf-8 -*-##### # Copyright (c), The AiiDA team.
The toytree ethos¶ Welcome to toytree, a minimalist tree manipulation and plotting library. Toytree combines a popular Tree data structure based on the ete3 library with modern plotting tools based on the toyplot plotting library. Introduction¶ The Newick format can be used for index construction in combination with the -A parameter. Names of files with sequences will be inferred from the names of leaves as [node_name].fa.
Overview ¶ Phylogenetic trees are the result of most evolutionary analyses. They represent the evolutionary relationships among a set of species or, in molecular biology, a … Introduction to Phylogenetics¶ Contents¶ Reasons for making a phylogenetic tree. Molecular phylogenetics
Questions: I’m trying to build a dendrogram using the children_ attribute provided by AgglomerativeClustering, but so far I’m out of luck. I can’t use scipy.cluster since agglomerative clustering provided in scipy lacks some options that are important to me (such as the option to specify the amount of clusters). I would be really grateful 27.11.2018 · Tools for working with the NCBI taxonomy database (and DIAMOND output) - 0.1.1 - a Python package on PyPI - Libraries.io
ete3 website ETE is a python programming toolkit that assists in the automated manipulation, analysis and visualization of phylogenetic trees. It provides a wide range of tree handling options, node annotation features, programmatic access to the phylomeDB database (containing thousands of pre-calculated phylogenetic trees), and automatic orthology and paralogy detection. 3. Installing ProPhyle¶ ProPhyle is written in Python, C and C++. It is distributed as a Python package and all C/C++ programs are compiled upon the first execution of the main program.
Python package for building, comparing, annotating, manipulating and visualising trees. It provides a comprehensive API and a collection of command line tools, including utilities to … Documentation - API Tutorial online or PDF. - API Reference guide - Command line tools online documentation - API code examples here. Talks. A brief introduction to ETE and its programmatic tree visualization features at the SciPy conference (2015) Courses.
ete3: The Environment for Tree Exploration (ETE) is a Python programming toolkit that assists in the recontruction, manipulation, Sphinx is a tool that makes it easy to create intelligent and beautiful documentation, written by Georg Brandl and licensed under the BSD license. I just discovered ETE3 yesterday and just tried quickly to run the following installation command using PIP on my win10 machine with a properly installed Python 3.6.3: python -m pip install --upgrade ete3 However, I am getting the following error: ##### Collecting ete3 Using cached ete3-3.0.0b35.tar.gz
Fire Emblem Heroes has a lot of things that beginners will miss. We've covered these things so you can progress fast. Fire emblem heroes beginner guide reddit Richmond 15.07.2018В В· For the last part i'll be going over how a newcomer can get themselves involved in the FEH community Useful links https://twitter.com/feheroes_news?lang=en h...
Introduction to Phylogenetics — Introduction to
Windowns Installation of ete3-3.0.0b35 with Python 3.6.3. 29.06.2019В В· Python package for building, comparing, annotating, manipulating and visualising trees. It provides a comprehensive API and a collection of command line tools, including utilities to work with the NCBI taxonomy tree. - etetoolkit/ete, The jkruppa/virDisco package contains the following man pages: aa_info_db aa_seqs artificial_genome_mapping build_index build_sample_info check_host collapse_df coverage_filter dna_seqs empty_map_pep_list ete2_plot fastq_quality_control fastq_trimmer fq2fa generate_decoy_reads generate_read get_bowtie2_cmd get_consensus_df get_pauda_hits get.
ete3 evol maximum number of taxa? - Google Groups
PythonQt download SourceForge.net. 3. Installing ProPhyle¶ ProPhyle is written in Python, C and C++. It is distributed as a Python package and all C/C++ programs are compiled upon the first execution of the main program., Documentation - API Tutorial online or PDF. - API Reference guide - Command line tools online documentation - API code examples here. Talks. A brief introduction to ETE and its programmatic tree visualization features at the SciPy conference (2015) Courses..
grimm.py¶ class bg.grimm.GRIMMReader ¶ Bases: object. Class providing a staticmethod based implementation of reading GRIMM formatted data file-like object and obtain a bg.breakp I just discovered ETE3 yesterday and just tried quickly to run the following installation command using PIP on my win10 machine with a properly installed Python 3.6.3: python -m pip install --upgrade ete3 However, I am getting the following error: ##### Collecting ete3 Using cached ete3-3.0.0b35.tar.gz
How do I get ETE3 to print a 7814 node sized tree. (alternate title: what is the best library to print out a 7814 node sized tree) I have a very very large reddit comment chain that I want a(n equally large) graphical representation of, and so far ETE3 has failed me. I am currently trying to use ete3 to test evolutionary hypotheses (codeml implemented in evol) on a set of candidate genes sequences in a large number taxa (>50 mammals from …
• Issue application to the BIS documentation and its application on the other similar projects • Issue identification from the NPP ETE3,4 tender process at purpose enhancement of the BIS documentation • Issue application to the BIS documentation and its application on the other similar projects Source code for aiida.cmdline.commands.work. # -*- coding: utf-8 -*-##### # Copyright (c), The AiiDA team.
def draw_children (node, node_label = None, show_pk = True, dist = 2, follow_links_of_type = None): """ Print an ASCII tree of the parents of the given node.:param node: The node to draw for:type node: :class:`aiida.orm.data.Data`:param node_label: The label to use for the nodes:type node_label: str:param show_pk: Show the PK of nodes alongside I am currently trying to use ete3 to test evolutionary hypotheses (codeml implemented in evol) on a set of candidate genes sequences in a large number taxa (>50 mammals from …
How do I get ETE3 to print a 7814 node sized tree. (alternate title: what is the best library to print out a 7814 node sized tree) I have a very very large reddit comment chain that I want a(n equally large) graphical representation of, and so far ETE3 has failed me. Conda mediated Installation¶ I’d really like to build a bioconda installation package, but would need some help. You can however install quite a few of the
*** By default, the R Studio session information will be stored in your home directory. If you are working with big data files, these files could be very big and fill up your home directory *** Dependencies¶ Funannotate has a lot of dependencies. However, it also comes with a few tools to help you get everything installed. The first is that of funannotate check.
Python package for building, comparing, annotating, manipulating and visualising trees. It provides a comprehensive API and a collection of command line tools, including utilities to … 16.07.2018 · I read the whole documentation on profile face heatmap generation but I could not find any way to add labels to the heatmap produced by ete3. For …
30.10.2019В В· Learn about Jupyter Notebooks and how you can use them to run your code. . Codecademy is the easiest way to learn how to code. It's interactive, fun, and you can do it with your friends. I just discovered ETE3 yesterday and just tried quickly to run the following installation command using PIP on my win10 machine with a properly installed Python 3.6.3: python -m pip install --upgrade ete3 However, I am getting the following error: ##### Collecting ete3 Using cached ete3-3.0.0b35.tar.gz
The jkruppa/virDisco package contains the following man pages: aa_info_db aa_seqs artificial_genome_mapping build_index build_sample_info check_host collapse_df coverage_filter dna_seqs empty_map_pep_list ete2_plot fastq_quality_control fastq_trimmer fq2fa generate_decoy_reads generate_read get_bowtie2_cmd get_consensus_df get_pauda_hits get Introduction¶ The Newick format can be used for index construction in combination with the -A parameter. Names of files with sequences will be inferred from the names of leaves as [node_name].fa.
16.07.2018 · I read the whole documentation on profile face heatmap generation but I could not find any way to add labels to the heatmap produced by ete3. For … grimm.py¶ class bg.grimm.GRIMMReader ¶ Bases: object. Class providing a staticmethod based implementation of reading GRIMM formatted data file-like object and obtain a bg.breakp
phyphy documentation — phyphy documentation
Plot dendrogram using sklearn.AgglomerativeClustering. Source code for gneiss.balances""" Balances (:mod:`gneiss.balances`) =====.. currentmodule:: gneiss.balances This module contains modules for calculating balances and creating ETE objects to visualize these balances on a tree., This will get you a list of ete3.TreeNode objects that represent the 5 most diverse possible strains. You can then use strainchoosr.get_leaf_names_from_nodes(diverse_strains) to get a list of names. Complete documentation on the strainchoosr API can be found at https://strainchoosr.readthedocs.io/api.html. Issues and Pull Requests.
PythonQt download SourceForge.net. def draw_children (node, node_label = None, show_pk = True, dist = 2, follow_links_of_type = None): """ Print an ASCII tree of the parents of the given node.:param node: The node to draw for:type node: :class:`aiida.orm.data.Data`:param node_label: The label to use for the nodes:type node_label: str:param show_pk: Show the PK of nodes alongside, ipyrad-analysis toolkit: abba-baba¶ The baba tool can be used to measure abba-baba statistics across many different hypotheses on a tree, to easily group individuals into populations for measuring abba-baba using allele frequencies, and to summarize or plot the results of many analyses..
OrthoEvolution — OrthoEvolution 0.9.0a2 documentation
BioHPC Cloud User Guide. I read the whole documentation on profile face heatmap generation but I could not find any way to add labels to the heatmap produced by ete3. For example in the following code, the 7 columns of the I read the whole documentation on profile face heatmap generation but I could not find any way to add labels to the heatmap produced by ete3. For example in the following code, the 7 columns of the.
Overview¶ This package focuses on inferring orthologs using NCBI’s blast, various sequence alignment strategies, and phylogenetics analyses including PAML, PhyML, ete3, and more tools. 30.10.2019 · Learn about Jupyter Notebooks and how you can use them to run your code. . Codecademy is the easiest way to learn how to code. It's interactive, fun, and you can do it with your friends.
def draw_children (node, node_label = None, show_pk = True, dist = 2, follow_links_of_type = None): """ Print an ASCII tree of the parents of the given node.:param node: The node to draw for:type node: :class:`aiida.orm.data.Data`:param node_label: The label to use for the nodes:type node_label: str:param show_pk: Show the PK of nodes alongside grimm.py¶ class bg.grimm.GRIMMReader ¶ Bases: object. Class providing a staticmethod based implementation of reading GRIMM formatted data file-like object and obtain a bg.breakp
Introduction to Phylogenetics¶ Contents¶ Reasons for making a phylogenetic tree. Molecular phylogenetics Conda mediated Installation¶ I’d really like to build a bioconda installation package, but would need some help. You can however install quite a few of the
ete3 website ETE is a python programming toolkit that assists in the automated manipulation, analysis and visualization of phylogenetic trees. It provides a wide range of tree handling options, node annotation features, programmatic access to the phylomeDB database (containing thousands of pre-calculated phylogenetic trees), and automatic orthology and paralogy detection. The toytree ethos¶ Welcome to toytree, a minimalist tree manipulation and plotting library. Toytree combines a popular Tree data structure based on the ete3 library with modern plotting tools based on the toyplot plotting library.
ete3 website ETE is a python programming toolkit that assists in the automated manipulation, analysis and visualization of phylogenetic trees. It provides a wide range of tree handling options, node annotation features, programmatic access to the phylomeDB database (containing thousands of pre-calculated phylogenetic trees), and automatic orthology and paralogy detection. • Issue application to the BIS documentation and its application on the other similar projects • Issue identification from the NPP ETE3,4 tender process at purpose enhancement of the BIS documentation • Issue application to the BIS documentation and its application on the other similar projects
GenBank Quality Control¶ Complete documentation lives at genbankqc.readthedocs.io. It is a work in progress. GenBankQC is an effort to address the quality control problem for public databases such as the National Center for Biotechnology Information’s GenBank. However, for power users, the TreeNode structure of toytrees provides a lot of additional functionality especially for doing scientific computation and research on trees. The TreeNode object in toytree is a modified fork of the TreeNode in ete3. Thus, you can read the very detailed ete documentation if you want a detailed understanding of the
grimm.py¶ class bg.grimm.GRIMMReader ¶ Bases: object. Class providing a staticmethod based implementation of reading GRIMM formatted data file-like object and obtain a bg.breakp Questions: I’m trying to build a dendrogram using the children_ attribute provided by AgglomerativeClustering, but so far I’m out of luck. I can’t use scipy.cluster since agglomerative clustering provided in scipy lacks some options that are important to me (such as the option to specify the amount of clusters). I would be really grateful
Overview ¶ Phylogenetic trees are the result of most evolutionary analyses. They represent the evolutionary relationships among a set of species or, in molecular biology, a … 16.07.2018 · I read the whole documentation on profile face heatmap generation but I could not find any way to add labels to the heatmap produced by ete3. For …
*** By default, the R Studio session information will be stored in your home directory. If you are working with big data files, these files could be very big and fill up your home directory *** Conda mediated Installation¶ I’d really like to build a bioconda installation package, but would need some help. You can however install quite a few of the
27.11.2018 · Tools for working with the NCBI taxonomy database (and DIAMOND output) - 0.1.1 - a Python package on PyPI - Libraries.io Dependencies¶ Funannotate has a lot of dependencies. However, it also comes with a few tools to help you get everything installed. The first is that of funannotate check.
Phylogenetic Course: Exercises¶ Basic phylogenetic tree making. We are going to use this to highlight some basic concepts and few exercises to learn tree making using some popular methods. Later we will use the workflows via ete toolkit version 3 or ete3 Source code for gneiss.balances""" Balances (:mod:`gneiss.balances`) =====.. currentmodule:: gneiss.balances This module contains modules for calculating balances and creating ETE objects to visualize these balances on a tree.